What are sequencing technologies?
Have you ever heard of the big keywords of sequencing techniques such as “Next Generation Sequencing”? And what about third generation sequencing? If your answer is not really well, you have definitely landed on the right page!
With this article you will learn more about the different sequencing technologies out there and understand their roles in current biomedical research right at the heart of the LUMC and in other research institutes across the world !
These big brand new technologies were first introduced to the medical world between 2005 and 2006. Starting from the first generation technologies – also known as Sanger sequencing – these sequencing technologies further developed into next generation sequencing (Illumina or MGI are some of the big names) and finally, into the most recent third generation technologies brought to us by the companies Oxford Nanopore and PacBio.
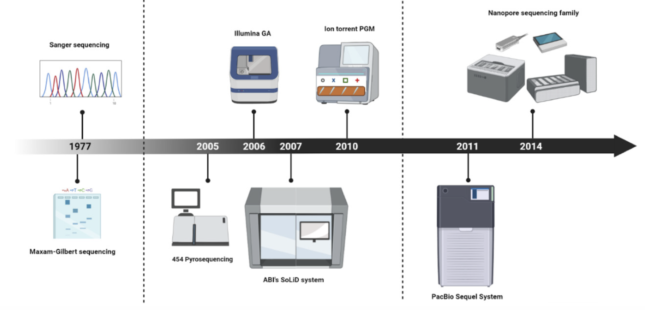
Method comparison
The main differences in these sequencing technologies lie in their capacity of reading strips of DNA of different lengths. In general, first generation technologies are capable of reading between short DNA fragments of around 100-600 base pairs per run, while next generation sequencing detects smaller reads from 50 to 600 base pairs at a time in a much more accurate way. Third generation sequencing can impressively generate over 10,000 base pair reads, which in other words means it can map over 10,000 DNA molecules!
Sequencing DNA fragments is truly important for uncovering genetic variants that are the cause of particular diseases in individuals. We can think of it as barcoding a certain variant that can cause a disease which can really help millions of individuals across the world.
Steps to sequencing
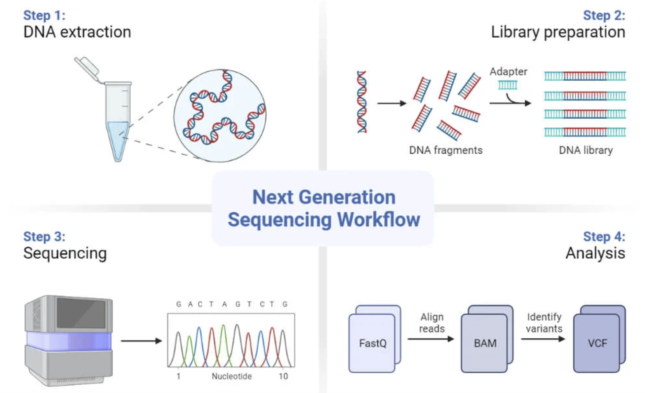
Lets dive deeper into how these technologies actually work: there are four main sequencing steps. First of all, the DNA samples need to be prepared and this is essentially where the DNA is extracted from a sample. In the course of this step, the DNA is purified of any proteins or transcription factors that might still be linked to it. During each of these steps, the scientist ensures quality control measures are taken to be able to trace back any mismatch with the results.
Secondly, a library needs to be prepared prior to the sequencing process. This is where we prepare our DNA so that it is compatible with the sequencer. This is done by fragmenting the DNA into smaller pieces onto which we add specific adapters to each end. The DNA is then amplified (multiplied) so that we can have many replicates of specific DNA regions; this will increase the accuracy of our final read of the DNA template after the sequencing has been performed.
The third step is the actual sequencing which occurs in cycles. During each cycle, one nucleotide at a time is going to be incorporated into the DNA which is building itself up again, just like we would with a lego tower. The strands that we had amplified in the previous step act as the blueprints that the DNA needs to know which nucleotides it needs to add in. Each nucleotide (A for arginine, C for cytosine, T for tyrosine and G for guanine) is tagged with a different color and has a different signal that can be detected by the device.
The final step is the analysis of the DNA reads which arrive in a FastQ file format. In summary, this step is where we reassemble the short reads of DNA together to get a bigger picture of what is happening. The post-processing consists of aligning the reads to a reference DNA genome, this is what information is contained in BAM files. Finally, once variants are identified, this information is stored into VCF files.
Short vs long read sequences
Interestingly enough, third generation sequencing works in a slightly different way than the first two sequencing methods. Instead of providing researchers with short reads of DNA, it provides long-reads of the DNA. This is extremely useful if we want to identify an event that is occurring that might for instance be linking multiple neighboring variants. We can imagine it as being able to zoom out on a variant and see if there are any regions that might be causing a certain disease as well. This really was a breakthrough, since the longer RNA strands get broken down, causing short reads, so to get long reads you need a much more delicate sequencer.
It is important to note that these technologies have as many advantages as they have disadvantages. While they provide researchers with extremely high throughput which means it can sequence many samples at the same time in a cost-efficient manner, their output can easily become very complex to analyze and requires sophisticated bioinformatics expertise to be interpreted. The most innovative machines have significantly reduced the amount of time they take for each run and have a much higher accuracy than the first technologies that were developed, but indeed, despite these advancements, there still is a large amount of error rates. The new rising challenges for these techniques is to increase the length of the reads per run as previously described. Obtaining longer reads of the DNA would allow for a more meticulous reassembly of genome sequences.
Relevance of these technologies
The way in which current research functions is that scientists are specialized in their area of expertise meaning they might learn to use one sequencing technology over another, for instance. Having this research focus can greatly aid analysis and diagnosis in particular disease such as cancer or neurological diseases. For example, short-read methods like Illumina are ideal to identify small genomes or targeted regions of the DNA. On the other hand, long-read technologies like PacBio or Oxford Nanopore sequencing are brilliant at unraveling some of the more complex structural variations in DNA regions. A combination of these different sequencing approaches can bring out a lot more information than what was initially found which can lead researchers to more comprehensive results and interpretations. In some cases, it could even enhance diagnosis of a rare disease!
Are you still curious about these technologies? Please go ahead and explore the resources below or get in touch with the Leiden Genome Technology Centre (LGTC) at the LUMC to learn more about these sequencing technologies!
Sources:
Aryal, S. (2022, August 5). Next-Generation Sequencing (NGS)- Definition, Types. Microbe Notes. https://microbenotes.com/next-generation-sequencing-ngs/
Cuber, P., Chooneea, D., Geeves, C., Salatino, S., Creedy, T. J., Griffin, C., Sivess, L., Barnes, I., Price, B., & Misra, R. (2023). Comparing the accuracy and efficiency of third generation sequencing technologies, Oxford Nanopore Technologies, and Pacific Biosciences, for DNA barcode sequencing applications. Ecological Genetics and Genomics, 28(100181), 100181. https://doi.org/10.1016/j.egg.2023.100181
NGS Workflow Steps. Illumina.Com https://emea.illumina.com/science/technology/next-generation-sequencing/beginners/ngs-workflow.html
How nanopore sequencing works. (n.d.). Oxford Nanopore Technologies https://nanoporetech.com/platform/technology
Leiden Genome Technology Center https://www.lumc.nl/en/research/facilities/leiden-genome-technology-center-lgtc/
Athanasopoulou, K., Boti, M. A., Adamopoulos, P. G., Skourou, P. C., & Scorilas, A. (2021). Third-generation sequencing: The spearhead towards the radical transformation of modern genomics. Life (Basel, Switzerland), 12(1), 30. https://doi.org/10.3390/life12010030
Lee, H., Gurtowski, J., Yoo, S., Nattestad, M., Marcus, S., Goodwin, S., Richard McCombie, W., & Schatz, M. C. (2016). Third-generation sequencing and the future of genomics. In bioRxiv. https://doi.org/10.1101/048603



0 Comments
Add a comment